
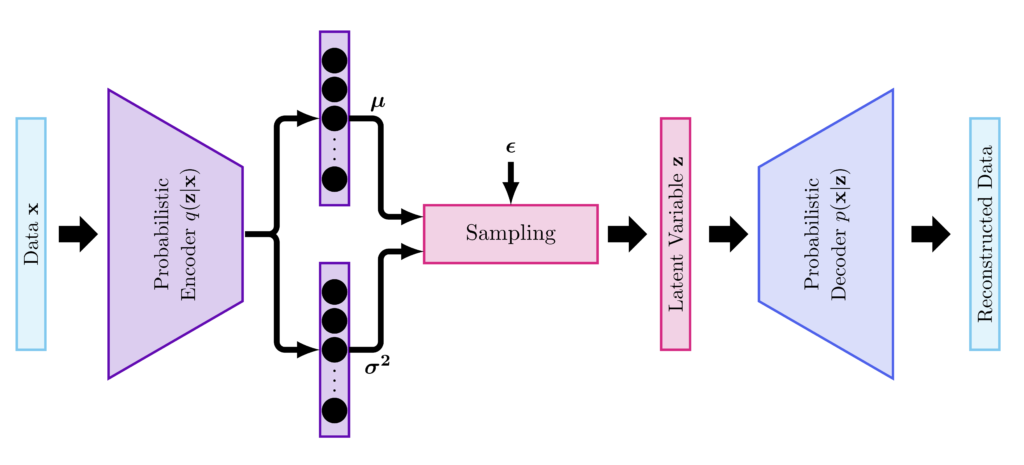
- The encoder encodes the inputs into a posterior probability distribution over the latent variables $\mathbf{z}$, $q(\mathbf{z}|\mathbf{x})$. $q(\mathbf{z}|\mathbf{x})$ can be chosen to be a Gaussian, as such the outputs of the encoder are mean, $\boldsymbol{\mu}$, and variance, $\boldsymbol{\sigma}^2$.
- A sampling step is required to sample the variables $\mathbf{z}$ from the estimated posterior density.
- The decoder decodes the latent variables $\mathbf{z}$ into a representation of the input data. In case of classification, for instance, the outputs are the class labels.
The dataset
We consider the MNIST dataset that consists of 28×28 grey level images of handwritten digits. The dataset has 60000 images for training and 10000 images for testing.

Code :
The encoder
We will code an encoder with convolutional layers. The outputs of the encoder will be fed to two fully connected layers to estimate the mean, $\boldsymbol{\mu}$, and variance, $\boldsymbol{\sigma}^2$, or more specifically $\log(\boldsymbol{\sigma}^2)$ to ensure the positivity of variance. The encoder requires setting the dimension of the latent variables $K$.
Code :
Sampling
In the code above there is a sampling step that consists of sampling the latent variables from the estimated distribution $q(\mathbf{z}|\mathbf{x})$. This step uses the reparametrization trick that ensures the backpropagation of the gradients. This step involves the formulation of $\mathbf{z}$ as function of $\boldsymbol{\epsilon}$.
Code :
The decoder
The decoder does not have to mirror the encoder, but I made it that way.
Code :
The loss function
The loss function writes, (for more details check here)
$$\mathcal{L}(\mathbf{x}) = \mathbb{E}_q\Big[\log\big(p(\mathbf{x}, \mathbf{z}) \big) – \log\big(q(\mathbf{z}| \mathbf{x})\big)\Big] = \mathbb{E}_q\Big[\log\big(p(\mathbf{x}|\mathbf{z}) \big)\Big] – D_{KL}\big(q(\mathbf{z}|\mathbf{x})||p(\mathbf{z})\big)$$
It consists of two terms.
- The first term depends on the data’s distribution. The MNIST dataset is binary (background in black and digits in white), accordingly it can be modeled by a Bernoulli distribution. In this case, $\mathbb{E}_q\Big[\log\big(p(\mathbf{x}|\mathbf{z}) \big)\Big]$ is given by the binary cross entropy available in Keras.
- The second term is the Kullback Leibler divergence between $q(\mathbf{z}|\mathbf{x})$ and $p(\mathbf{z})$, which are both Gaussians. We assume that $p(\mathbf{z}) = \mathcal{N}\left(\mathbf{0}, \mathbf{I}\right)$, accordingly,
$$D_{KL}\big(q(\mathbf{z}|\mathbf{x})||p(\mathbf{z})\big) = \frac{1}{2} \sum_{k=1}^K \Big( -1 + \boldsymbol{\mu}^2_k + \boldsymbol{\sigma}^2_k – \log\big(\boldsymbol{\sigma}^2_k\big)\Big)$$
Putting everything together!
We combine the encoder and decoder together.
Code :
Let's run the code!
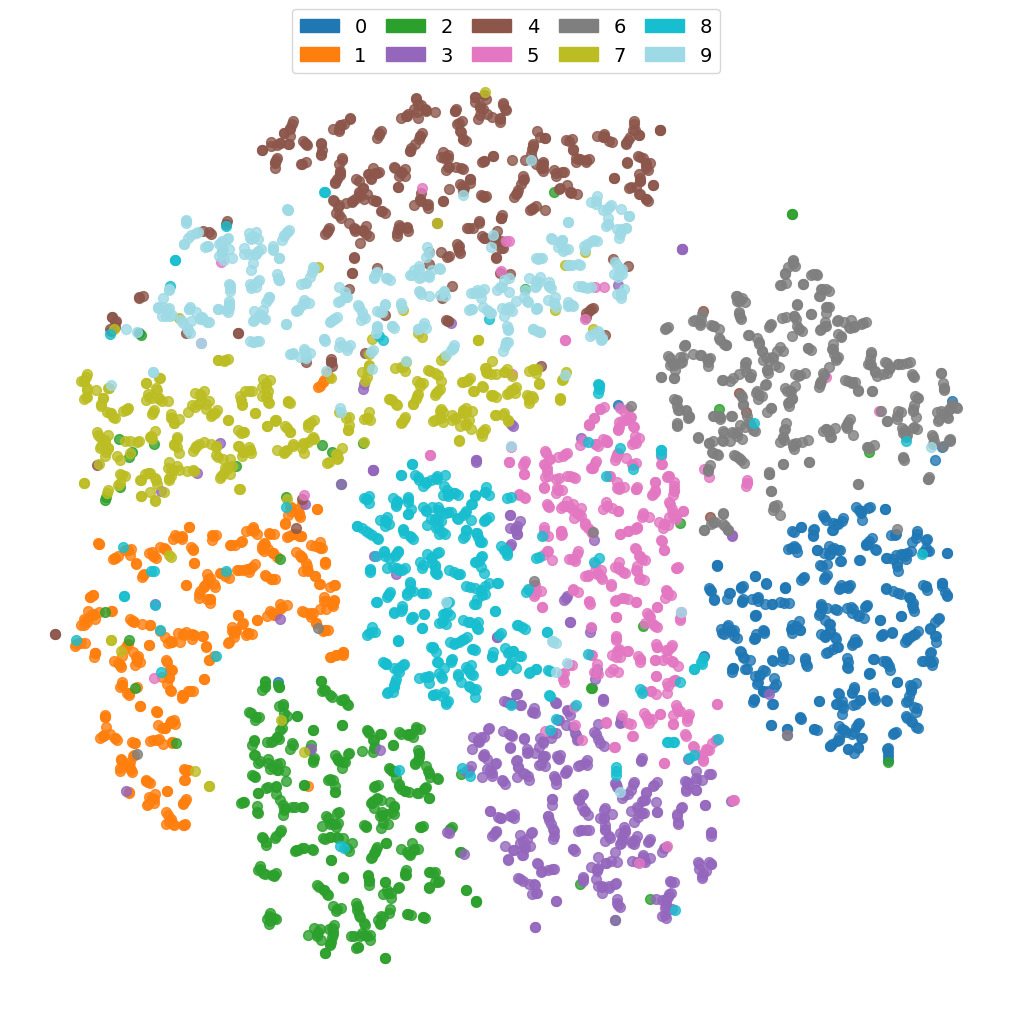
Below we show the t-SNE (t-distributed Stochastic Neighbor Embedding) of the original dataset MNIST and the corresponding embeddings $\mathbf{z}$. Note how the embeddings improve the classes’ separation.

Original digits and reconstructed digits by the implemented VAE


Was this post useful? Let me know what do you think about it below…